2011 Senior Thesis Abstracts
(CLASS OF 2011: If your thesis abstract is not currently included on this page and you would like it to be, please follow this link.)
Replication of Barley yellow dwarf virus in Saccharomyces cerevisiae
Kaileigh Djanaba Ahlquist
(Advisor: Russell)
All known cellular life on Earth serves as a host for viruses. Disease caused by viruses is frequently devastating, leading to the loss of health or life in the host. Yet, while viruses are frequently destructive to cellular life, viruses are obligate intracellular parasites, depending on their hosts to produce many of the products they require for their life cycles. Understanding how the host allows and facilitates viral infection is important because it could allow the development of anti-viral drugs, or point out ways to modify hosts so that host-machinery required by viruses would no longer be accessible. Host factors required for viral replication can frequently be discovered through genetic manipulation of the host, however, this is only possible in hosts with available genetic tools and techniques. When the natural host of a virus does not have the necessary genetic resources available, model organisms can play important roles as heterologous hosts, allowing for the study of the virus in the context of a malleable and well-understood host organism. Recently, the use of yeast as a heterologous host for the discovery of host factors has become an important technique in the study of the (+) sense ssRNA plant viruses Tomato bushy stunt virus and Brome mosaic virus. This thesis attempted to bring this technique to bear on one of the most important and influential plant viruses, Barley yellow dwarf virus (BYDV). Prior to this thesis, a modified BYDV gRNA, known as the mini-genome replicon (MGR) was constructed, expressing a reporter gene as an indication of MGR replication in yeast. Due to the low and variable expression of the reporter gene in version of the MGR, this thesis created a modified MGR, replacing the reporter gene with a selectable marker. A plasmid bearing the modified MGR was transformed into yeast, along with two plasmids expressing proteins necessary for the replication of the BYDV gRNA. Expression of the selectable marker from the MGR was too low in initial experiments to allow for growth on selective media in this thesis. While these early results are inconclusive, it is possible that further screening of yeast containing the modified MGR will provide a yeast strain maintaining robust replication suitable to study BYDV replication in the future.
The Effect of PinX1 fusion protein on telomerase activity of Xenopus laevis in vitro
Giancarlo N. Bruni
(Advisor: Shampay)
Telomeres are necessary for chromosome stability and cellular survival. Telomerase extends the telomere, preserving telomere length. Shelterin, a protective protein complex that is located at telomeres allows telomerase access to the telomere and protects the telomere from damage. PinX1 has been shown to interact with a component of Shelterin and has an inhibitory effect on telomerase activity. Xenopus have been shown to have telomere sequence and structural similarity to a number of vertebrates, including humans. In Xenopus, unlike humans where expression is confined to germline and proliferative cell compartments, telomerase is constitutively expressed in somatic tissues. Thus, telomerase may be regulated differently in Xenopus than in humans. Xenopus laevis PinX1 has been shown to have a dose dependent inhibitory effect in a telomerase assay. The current research attempts to replicate the inhibitory effects of PinX1 on telomerase activity. Two homologs, Human and Xenopus, were expressed separately in E. coli and then purified. These proteins were added to an in vitro telomerase assay to observe their effect on telomerase product relative to positive and vector controls. Results indicated that with increased PinX1 concentration, telomerase product diminished. PCR activity also decreased with high PinX1 concentration and thus the results were inconclusive. The direct inhibitory effect of PinX1 on telomerase activity could not be established.
The role of non-floral pigments and environmental plant stress in the divergence of two Delphinium species
Charlene M. Grahn
(Advisor: Karoly)
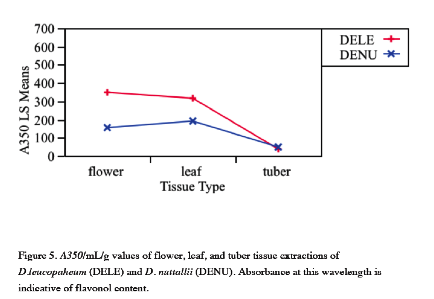
Differences in flower color between two recently diverged plant species, Dephinium leucophaeum and Delphinium nuttallii, are caused by differing expression of flavonols and anthocyanins within their floral tissue. Anthocyanins and flavonols also function as antioxidants within plant tissues, absorbing reactive oxygen species generated when plants are under stressful conditions, such as drought, flooding, and temperature extremes. If different levels of anthocyanins and flavonols are expressed in non-floral tissues between the two species, it may cause a difference in the ability of the two species to cope with environmental stress. It had been observed that D. leucophaeum was generally found in flooded conditions while D. nuttallii was not, and it was hypothesized that a pleiotropic effect may be causing the tubers of D. leucophaeum to express high levels of flavonols just as in the species' floral tissue. This would in turn lead to geographic isolation and explain the mechanism behind their speciation. In this study, floral, leaf, and tuber tissue of D. leucophaeum and D. nuttallii were examined for differences in anthocyanin and flavonol expression through the use of pigment analysis. It was found that flavonol levels in D. leucophaeum leaves and flowers were significantly higher than those in D. nuttallii. Flavonol levels in tubers of both species were found to be significantly lower than leaves and flowers of either species, and no difference in flavonol level between the two species' tubers was observed. It was also found that the leaves of D. leucophaeum were richer in anthocyanins than D. nuttallii leaves. No significant difference was observed in the flower or tuber tissue between species. An attempt was made to measure the antioxidant capacities of the different tissues using the oxygen radical absorbance capacity (ORAC) assay, which was performed on a real-time qPCR machine. The use of the assay for this urpose was ultimately unsuccessful. The low levels of flavonols and anthocyanins found present in the tuber tissue by the pigment assay makes the initial hypothesis of tuber flavonol levels impacting flooding resistance seem unlikely.
Variation in Developmental Patterns in Two Geographically Separated Populations of Bombina orientalis in the Republic of Korea
Advait Mahesh Jukar
(Advisor: Kaplan)
This thesis used a common garden experiment, conducted in the field in the Republic of Korea, to investigate the effect of the interaction between variable levels of maternal investment and different levels of diel thermal fluctuations on the early development of two populations of Bombina orientalis in order to determine whether the two populations are locally adapted to their native habitats. In addition, in order to understand the patterns of morphological variation induced by diel thermal fluctuations, a laboratory study was modelled to predict field responses from responses in constant laboratory environments. The common garden experiment revealed that maternal investment was positively correlated with total length, tail length, snout-vent length (SVL) and the ratio of tail length to SVL while thermal variance was negatively correlated with total length, tail length and the ratio of tail length to SVL. These relationships were consistent at the hatching stage and 24 hours after hatching. Significant population level differences were also observed for total length, tail length and the ratio but these effects were modified by egg size and thermal variance. Using total length and the ratio of tail length to SVL as surrogates of fitness, the experiment suggested that large JungBong eggs produced the largest and fastest hatchlings in the low thermal variance environment while large ChunCheon eggs produced the largest and fastest hatchlings in the high thermal variance environment. Since the low thermal variance and high thermal variance environments were modelled after the JungBong and ChunCheon environments respectively, the results of the experiment suggest that the two populations may be locally adapted. The laboratory model for development at different temperature ranges did not match with the field results suggesting that certain morphological traits might be buffered against thermal variance or there might be an interaction between mean temperatures and thermal variance. This study helps answer questions about rapid adaptive evolution in new environments and expands on the role of environmental factors as inducers of phenotypic variation and agents of natural selection.
Hybe and go Seq: application of genomic tools to sociogenomic questions in the cichlid genus Julidochromis
Quinn Langdon
(Advisor: Renn)
Through developments in sequencing technology, new questions in non-model organisms can be addressed. Genomic influence on interesting social behavior can be addressed though the field of "sociogenomics". This thesis works with two species from the genus Julidochromis of cichlid fish. The species J. transcriptus is sex-role conventional, where in natural pairs the male is larger and more aggressive while the female is more submissive and parental. Conversely, in the species J. marlieri this is reversed, and the female is dominant while the male is submissive. This is an excellent systems in which to address the genetic control of these stereotypical social phenotypes. I will address the two common ways to assay whole genome expression; microarrays and RNA-Seq. I am basing my thesis on previous cDNA microarray work that analyzed patters of gene expression within species. I have completed cross-species hybridizations and comparisons that provide greater power in determining gene expression differences between social behavioral phenotypes. In order to complete cross-species hybridizations for expression, genomic sequence differences between the species must be taken into account. I have identified 8 genes that are significantly diverged between my species. These eight genes bias expression results and must be removed from expression analyses. I have further complete direct between species microarray comparisons of gene expression to identify patterns that are associated with sex, species, and behavioral sexrole. A module of 58 genes that are potentially associated with female specific aggression was identified. Furthermore, I address how the emerging method of RNA-Seq will enhance our understanding of genomic control of these social behaviors. I also explain downsides of RNA-Seq. Techniques for acquiring and analyzing RNA-Seq data are discussed. Current microarray work and future RNA-Seq work will add to the growing sociogenomics literature on genomic and evolutionary control of social behaviors.

Photographs of J. transcriptus (left) and J. marlieri (right)
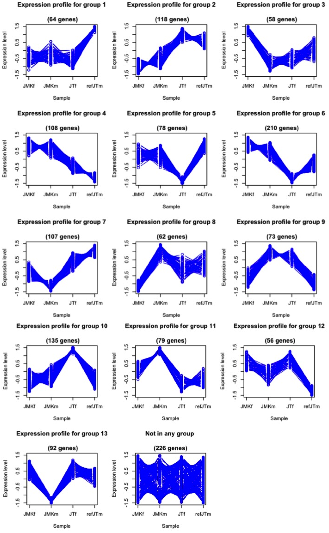
Figure 26. Clustering of microarray expression results Kmeans clustering was applied to all the significantly differently regulated genes to produce groups of similar expression across the phenotypes. Expression level is expression ratio. JMKf= J. marlieri females, JMKm = J. marlieri males, JTf = J. transcriptus females, and refJTm= J. transcriptus males.
An Investigation into the Secretion and Processing of GFP-tagged ELH Prohormone from Transgenic Bag Cells
Tyrone Lee
(Advisor: Arch)
Neuroendocrine systems mediate multiple aspects of physiology and behavior. The secretory pathway is a vital process in neuroendocrine cells, yet the cellular mechanisms of secretion are not well understood. Here we probe the well-characterized hormonal model, the reproductive bag cell neuroendocrine system of the sea slug Aplysia californica. The bag cells of Aplysia mediate egg-laying behavior through the coordinated secretion of a suite of peptides derived from a single gene product, the egg-laying prohormone (proELH). A previous study had produced a chimeric vector fusing proELH with a traceable tag called green fluorescent protein (GFP). It was anticipated that when this vector is transfected into bag cells, the mature protein can be reliably traced by florescent microscopy. However not only have individual bag cells proven difficult to culture for microscopy, the fusion protein, proELH-GFP, is not processed into a mature peptide within the transfected cells making it less desirable as a tracker of ELH. It was unknown if this product was even capable of being secreted. The goal of this project was to develop a reliable method of retrieving proteins from the incubation medium in cultured bag cell organs and then screen for GFP-tagged proteins by SDS-PAGE and western blots. So far, GFP-tagged proteins are detected from precipitated medium. However, results have been obscured by non-specific binding by anti-GFP antibody and uncertain rates of transfection. Better detection methods must be developed in order for better characterization of proELH-GFP.
Telomeric localization of shelterin protein xTRF1 in somatic Xenopus laevis cells
Sophie E. Mayer
(Advisor: Shampay)
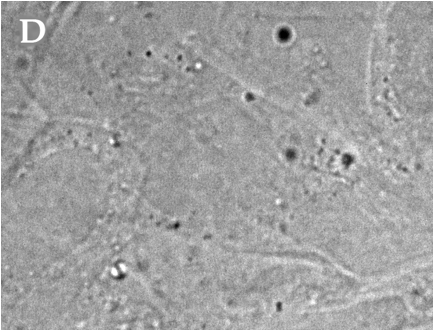
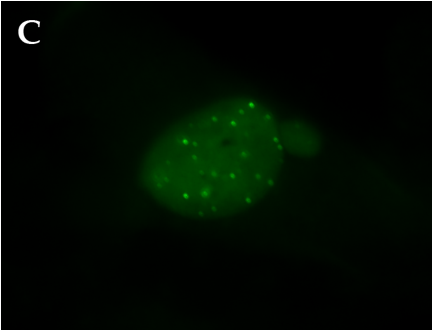
Telomeres are specialized terminal tracts of chromatin that protect eukaryotic chromosomes from degradation and inappropriate DNA repair and extension. Telomeric DNA is bound by a collection of shelterin proteins involved in regulating telomere maintenance; in humans, Telomere Repeat-binding Factor 1 (TRF1) is one of the proteins that anchors the shelterin complex. DNA replication requires that chromosome ends be made accessible to processing by polymerases, endonucleases, and in some organisms and cell types, terminal transferases. Cell cycle-dependent regulation of the shelterin proteins is thought to mediate telomere accessibility. Though humans and Xenopus laevis express homologous TRF1 proteins, X. laevis TRF1 (xTRF1) appears to be bound for only a minimal portion of the cell cycle in cultured egg extracts. The following work seeks to characterize TRF1 binding and expression in vivo in a somatic Xenopus cell line. Cells were transfected with a fluorescently tagged xTRF1 construct, and fusion protein localization observed by microscopy. xTRF1 displayed a punctate nuclear distribution pattern characteristic of telomeric localization in interphase cells, indicating that telomere regulation in X. laevis is not dissimilar to that of human systems, despite earlier evidence to the contrary.
The relation of xTERT transcript abundance to relative telomerase activity in adult Xenopus laevis tissues
Ella Stern
(Advisor: Shampay)
Telomeres are the ends of linear chromosomes that protect vital genetic information from being lost during DNA replication. Telomeres need to maintain a certain length in or- der for cells to continue dividing; the ribonucleoprotein telomerase composed of a reverse transcriptase and an RNA template, extends telomeres to maintain this length. Though telomerase is inactive in most somatic human tissues, its activity is widespread in Xeno-pus laevis somatic tissues, raising the question of how telomerase is regulated in Xenopus.
The transcript level of TERT, the gene encoding for the reverse transcriptase component of telomerase, has been found to directly correlate with telomerase activity in human cancer cells and proliferative and developing tissues. This experiment explored whether there is a similar relationship between TERT transcription and telomerase activity in Xenopus, in which telomerase activity is only relatively different from tissue to tissue. Quantitative PCR was used to analyze relative transcript abundances in tissues recorded to have either high or low telomerase activity from three X. laevis individuals. More abundant TERT transcript in low telomerase tissues indicate that transcriptional regulation of TERT is not related to telomerase activity in Xenopus as it is in humans, and some other form of regulation may be occurring. These preliminary results should be further explored with data from more individuals, as well the use of a more reliable normalizing gene.

Image: A diagram (not to scale) of the sequenced xTERT gene adapted from (Kuramoto et al., 2001), with the open reading frame boxed in. Regions v-I-v-IV conserved in vertebrates are colored red. The separate sections of the reverse transcription motif (RT motif) are labeled, along with the T motif. The red line denotes the part of the gene that will be ampliï¬ed in qPCR in this thesis. Nucleotide numbers are listed at the beginning and end of the transcript, either side of the open reading frame, as well as at the portion to be ampliï¬ed during qPCR.
Anti-estrogen effects of melatonin on gene regulation and MAPK signaling in MCF-7 human breast cancer cells
David E. Toffey
(Advisor: McClellan)
Melatonin is widely known for its role in communicating photoperiod length to the vertebrate circadian clock. Increasingly, melatonin is recognized as a possible oncostatic agent for a variety of human malignancies. In humans, there is growing acceptance that light at night interrupts endogenous secretion of melatonin and that this interruption is correlated to increased risk of cancer. I investigated the antiproliferative effects of melatonin on asynchronous populations of dividing estrogen-responsive MCF-7 human breast cancer cells. Using a fluorescence-based cell proliferation assay and PAGE/western analysis employing phosphospecific antibodies, I found that combinations of melatonin and estrogen act synergistically to inhibit cell growth, and this effect is mediated by inhibition of the phosphorylation of the so-called extracellular regulated kinase (ERK) pathway. Additionally, melatonin appeared to reduce estrogen-stimulated expression of the cell cycle regulatory gene, cyclin D1. The data support the idea that melatonin is oncostatic. Furthermore, the results suggest melatonin may, over long-term exposure, indirectly interfere with the action of cellular growth signals, leading to decreased mitogen activation in dividing cells.
Evolving Dispersal and Survival: Simulating Eco-Evolutionary Dynamics in Rana aurora
Ross Young
(Advisor: Kaplan)
Recent theoretical and field work has advanced the study of "eco-evolutionary dynamics," the concept that not only can ecological dynamics influence evolutionary change, but rapid evolutionary change can also feed back into ecological dynamics. Understanding these feedbacks could have a great impact on the study of ecology and evolution, as well as applied conservation disciplines. In this thesis, I use a new simulation program, Nemo, to examine the eco-evolutionary dynamics of dispersal. These simulations were parameterized with data gathered on the northern red-legged frog (Rana aurora), a widely dispersing Pacific-Northwestern frog threatened by human activity. From my simulation results, I address three hypotheses: first, that mortality during dispersal and/or environmental stochasticity will affect metapopulation persistence, second, that mortality during dispersal and environmental stochasticity will result in rapid evolution of the dispersal trait, and third, that evolution of dispersal will affect metapopulation persistence. Statistical analysis of my simulation results provided strong support for all three of these hypotheses, which I interpret in the context of Rana aurora conservation. My goals with this work were threefold: to examine the eco-evolutionary interactions between evolution of dispersal and metapopulation persistence, to assess Nemo as a research tool, and to apply my results to the conservation of Rana aurora.