Research
Our long-term research goal is to study the molecular mechanisms of how enteropathogenic E. coli (EPEC) causes disease. We have two main projects focusing on EPEC virulence gene regulation. The first area of study concerns the regulation of EPEC virulence factors, and mechanisms of control by a novel protein called Ler. Funding for this set of projects has included a three-year, NIH AREA grant (2R15AI47802-03).
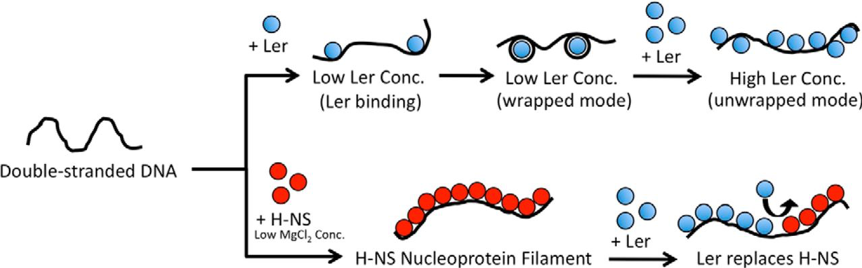
Models of Ler binding to DNA and its role in gene regulation. At nonsaturated condition or low tension, Ler can bind DNA in the wrapped mode. At a higher concentration (Conc.) of Ler or high tension, Ler binds DNA in an unwrapped mode in a largely noncooperative process to form Ler·DNA nucleoprotein complex array. H-NS, however, forms rigid nucleoprotein filament at low MgCl2 concentrations (<5 mm) through a cooperative binding process. The formation of nucleoprotein filament may silence genes by blocking RNA polymerases from accessing the promoters or blocking translocation of RNA polymerases. Ler can replace H-NS nucleoprotein filaments in some environmental conditions (such as low KCl concentration). This replacement of H-NS by Ler, together with Ler's lack of cooperativity, may serve as the basis to understand Ler's anti-silencing activity. From Winardhi et al. (2014) Journal of Biological Chemistry 289:13739-13750.
A second project involves delineating the regulatory network controlling the initial stages of EPEC infection. The plasmid-encoded regulator, PerC, in addition to ler, controls a number of genes involved in niche adaptation. Thus, we are interested in how PerC confers a predicted selective advantage to the bacterium at the site of infection- the small intestine, and the molecular mechanism by which this small regulatory protein controls gene expression. Funding for this project has included an award from the M.J. Murdock Charitable Trust and an NIH Exploratory/Developmental Research Award (1R21AI115193-01).
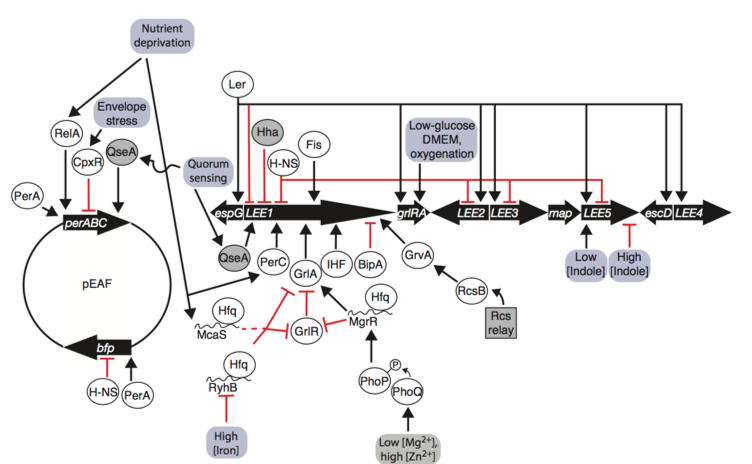
EPEC LEE and pEAF gene regulation is influenced by environmental inputs. The LEE1 operon encodes Ler, which promotes transcription of all operons and auto-represses its own promoter. LEE1 is regulated by a number of nucleoid associated proteins (Ler, H-NS, Hha, Fis, and IHF), pEAF-encoded regulator PerC, LEE-encoded GrlA/GrlR, quorum-sensing factor QseA, BipA, and GrvA. RelA, CpxR, and QseA regulate the per operon on the pEAF, PerA promotes bfp transcription and auto-activates per.Environmental signals are transmitted through phosphorelay systems, sRNAs, stringent responses, quorum-sensing responses, and envelope stress responses to affect transcription of the LEE and pEAF. From Mellies et al. (2018) Frontiers in Cellular and Infection Microbiology. https://doi.org/10.3389/fcimb.2017.00032.
A third project in the laboratory focuses on isolating and characterizing environmental bacterial isolates that can degrade commercial plastics. With the growing problem of plastic pollution, both on land and in our oceans, we see the biodegradation of these waste products to be an important part of the solution to this global issue. For her senior thesis Morgan Vague (Reed ’18) isolated three Pseudomonads that degrade polyethylene terephthalate, or PET plastic, the material used for disposable water bottles. We are looking to devise pre-treatment strategies to make the degradation more rapid, and to gain a better understanding of the related biological and chemical processes. We are currently seeking funding for this exciting set of projects.
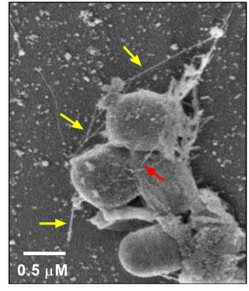
Pseudomonas morganensis and Bacillus cereus forming a biofilm on PET plastic. PET strips were incubated in carbon-free media inoculated with lipase positive bacteria, and imaged with scanning electron microscopy. Pili formation by bacteria aid in adherence and biofilm formation. Yellow arrows denote pili attached to PET plastic while red arrow denotes a pilus between bacteria, aiding in cell-cell adhesion (Morgan Vague, Senior Thesis, 2018). Image courtesy of Claudia S. López, PhD, Director Multiscale Microscopy Core at OHSU.
Lab Folks 2017/18

Research Associates: Amy Platenkamp, Morgan Vague
Thesis Students: Eliotte Garling, Katie McPherson, Jethary Rader, and Trevor Soucy
Independent Study: Colin Hawkinson