Renn Lab Thesis Project
2011 Reed Graduate
SEAN MADEN
Small Fish in a Small Pond: Observed Declines in Genetic Diversity Across Successive Generations of A Captive Astatotilapia Burtoni Lineage, Using Microsatellite Markers
This thesis studies the extent to which genetic diversity in a population of the South African cichlid fish species, Astatotilapia burtoni has been affected by a laboratory environment. Recent literature discusses the importance of maintaining genetic variation for captive populations to increase the chances of successful reintroduction into natural environments, but the problem of maintaining natural genetic variation in laboratory populations for the purpose of research is less widely addressed. The latter problem served as the primary motivation for this thesis research.
Genetic variation was assessed temporally across three generations in the test population. Three sample groups were taken from the parent generation at three distinct times (2004, 2006, and 2007), and also from the first and second (F1 and F2) generations bred in the laboratory.
Microsatellite markers were used to assess variation in each of the five sample groups. Screening is essential in selecting loci for use in population genetics studies, as ideal markers in these studies are loci which are subject to negligible selective pressure, called neutral markers. For microsatellites, selection is generally assumed to reduce variation, so loci were evaluated for high levels of variation using a library of primer sets established in tilapia.
Allele variation at microsatellite loci tends to consist in additions or subtractions of short (1-6bp) nucleotide repeats (Fig. A shows the mechanism by which this occurs). Using high-density agarose gel electrophoresis, four microsatellite loci associated with three linkage groups were selected for further study (Lee et al 2005). Three of these loci showed high allele size variation, and the fourth showed high specificity for targeted microsatellite regions.
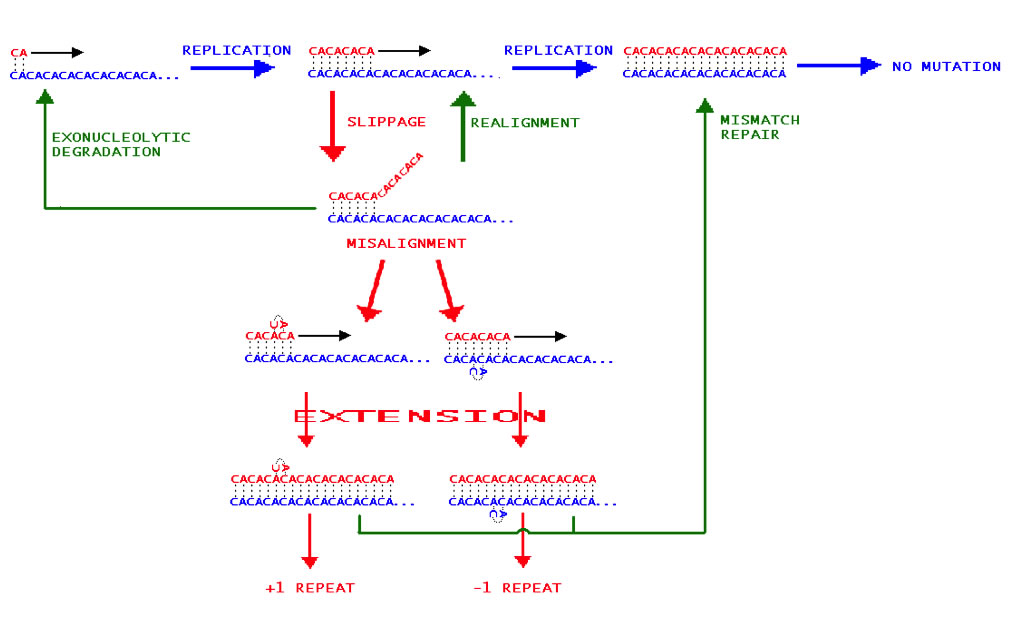 |
| Fig. A – The Slip Strand Mispairing mechanism for microsatellite repeat addition/subtraction during DNA replication. The example microsatellite region consists in dinucleotide CA repeats (blue strand is the DNA template, red is the complementary strand). Blue arrows and text show normal DNA replication stages, red arrows and text show errors during replication, and green arrows and text show normal repair mechanisms. |
Fluorescently-labeled forward primers targeting the four target loci were obtained and paired with reverse primers, and over twenty individuals from each of the five sample groups was analyzed by capillary electrophoresis in Fragment Analysis, to precisely determine allele fragment sizes at each locus (in bp, OHSU Core lab).
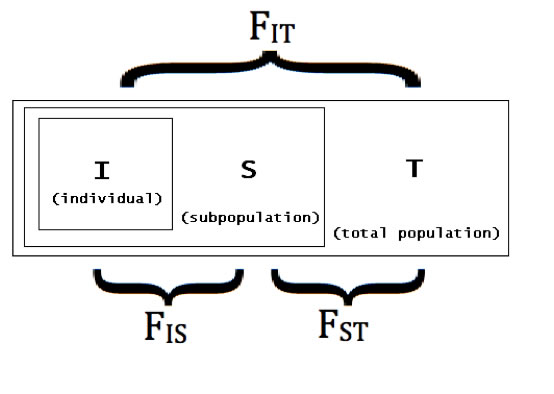
Allele size data were used to perform statistical analyses with three of the four target loci (the fourth was excluded based on low fragment size variation, which suggested it would not be informative as a marker). Genetic diversity was assessed by calculation of inter and intra-generational parameter estimates, including F-statistics (see Fig. B).
|
Fig. B – Schematic of F-statistics for a subdivided population. F-statistics reflect the probability that two paired alleles are identical by descent, as indicated across the three distances IS (for an individual in a subdivided population), ST (averaged for individuals in subpopulations, across the entire grouping of subpopulations), and IT (for an individual in the entire grouping of all subpopulations). For any of these parameters, a calculation of "0" indicates the population is entirely outbred, while a population with parameter "1" is entirely inbred. |
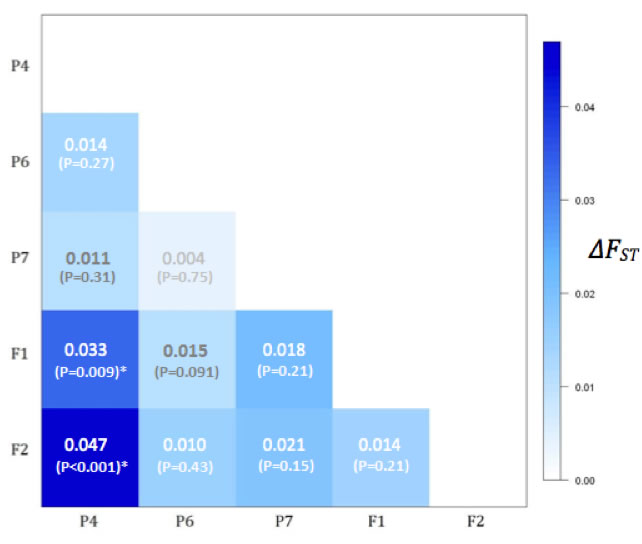
Results indicate substantial loss of genetic variation, including significant departures from Hardy-Weinberg Equilibrium expectations across all sample groups, and significant genetic divergence between generations (Fig. C depicts comparisons of FST parameters between P4 and the four other sample groups). A further test calculating the M coefficient implicates bottleneck-like phenomena in contributing to losses of genetic diversity (Garza and Williamson 2002). These results lay a foundation for further work investigating genetic diversity in laboratory environments, and could be used as the basis for a breeding protocol that better maintains genetic variation at levels observed for populations in natural environments.
|
Fig. C - Inter-population pairwise comparisons of FST statistics. The only comparisons showing significant differences are between P4-F1 (?FST = 0.03; P < 0.001) and P4-F2 (?FST = 0.05; P < 0.001). Numbers in the matrix are ?FST and P-values. |
Works Cited:
Garza, J, E Williamson. Detection of reduction in population size using data from microsatellite loci. Molecular Ecology, (10):305-318, 2001.
Lee, BY, WJ Lee, JT Streelman, KL Carleton, AE Howe, G Hulata, A Slettan, JE Stern, Y Terai, and TD Kocher. A second generation genetic linkage map of tilapia (Oreochromis spp.). Genetics, 170(1):237-244, May 2005.
This research was supported in part by a grant from the Reed College Biology Undergraduate Research Project Program (B.U.R.P.)

